 What are the inter-protein linkers of PKSs?
What are the inter-protein linkers of PKSs?
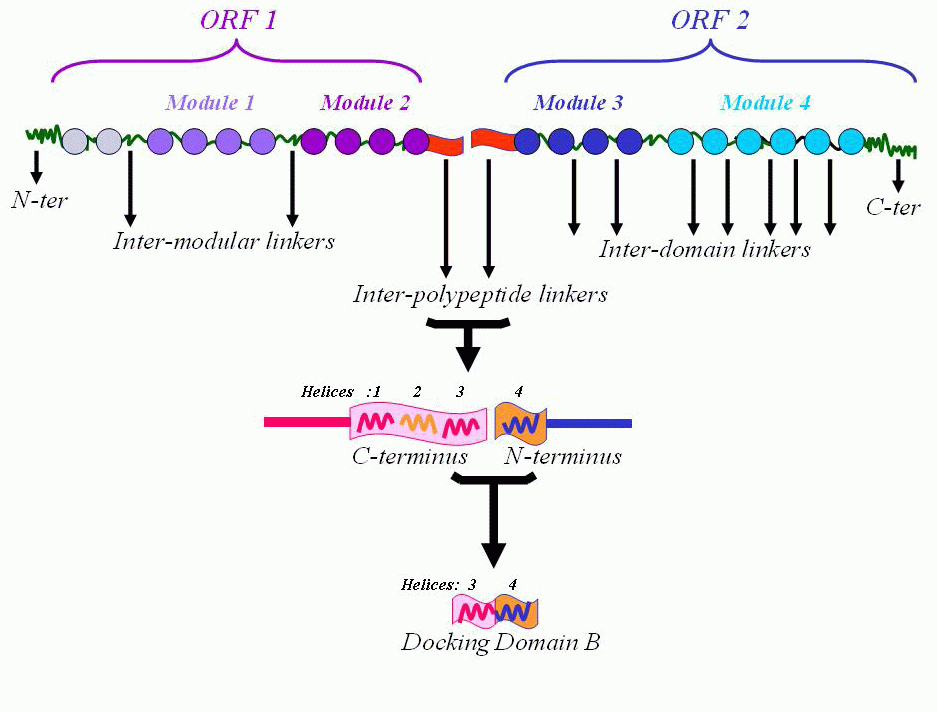
The domains of modular PKSs are independent globular entities separated by polypeptide segments referred to as the ‘inter domain linkers’ while the linkers that connect an upstream module to the downstream module are termed the ‘inter modular linkers’. Apart from these two categories, linkers are present at the termini of PKS ORFs and these are referred to as the ‘inter polypeptide linkers’. These have also been termed as the terminal docking domains of PKSs.
Thus, inter polypeptide linkers comprise of sequences at the termini of the ORFs, and are very short stretches of amino acids. These have been extensively studied and reported to play a significant role in inter modular cross-talk between the consecutive ORFs. The C-terminal (Cter) linker is speculated to specifically pair with the N-terminal (Nter) linker region of the succeeding ORF in modular clusters, thereby making cognate interacting partners of Nter-Cter linkers.
Protein-protein interactions between terminal docking domains of successive multienzymes promote their correct positioning within the assembly line.
 How do docking domains interact?
How do docking domains interact?
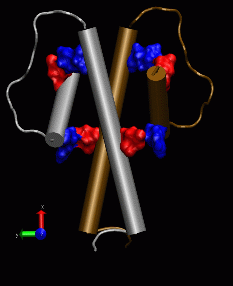
The key interactions between the DEBS2-3 cognate pair of docking domains have been identified through the NMR solution structure of a 120 residue polypeptide representing a typical pair of such domains, fused at their respective C and N termini. They adopt a stable dimeric structure which reveals the detailed role of these (predominantly helical) domains in docking and dimerization by modular polyketide synthases. (Broadhurst et al, 2003).
The first 60 residues form the docking domain A while the consecutive 60 (consisting of C-terminus and N-ternminus)
form the actual docking interaction, and is called the docking domain B.
These domains recognise their cognate partners via electrostatic interactions, which exist at the junctions of spatially close helical stretches within the docking domain B.
 How is linker analysis done at this site?
How is linker analysis done at this site?
The docking domain structure provides evidence that the specificity between upstream and downstream PKS linkers is responsible for the efficiency of chain transfer and the maintenance of the correct order of ORFs in a modular cluster.
We have scanned for the reported ionic pairs between adjacent and non adjacent modules of several characterized modular PKS clusters, in order to investigate whether the docking code can be used to predict the functional order of substrate channeling in a modular PKS cluster.
The prediction of the correct order of substrate channeling is essential for in silico identification of polyketide products of uncharacterized modular PKS clusters. Therefore we have attempted to investigate if there is a PKS docking code which could help to decipher the correct order of substrate channeling in an uncharacterized PKS cluster.
By highlighting how substrate channeling can allow PKS modules to effectively accept and process non cognate substrates, these studies provide a rational basis for examining the enormous untapped potential for combinatorial biosynthesis via module rearrangement