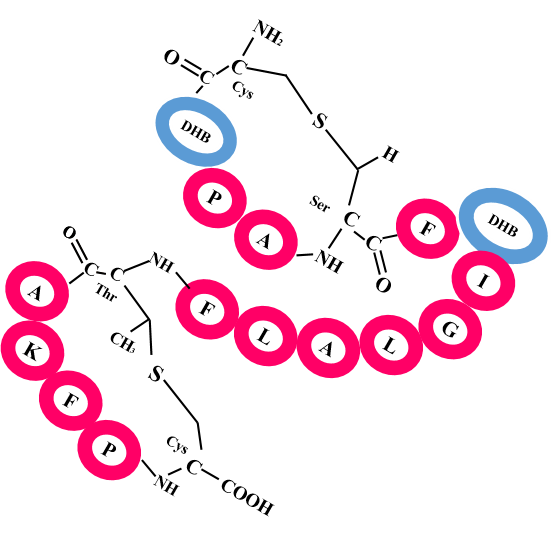
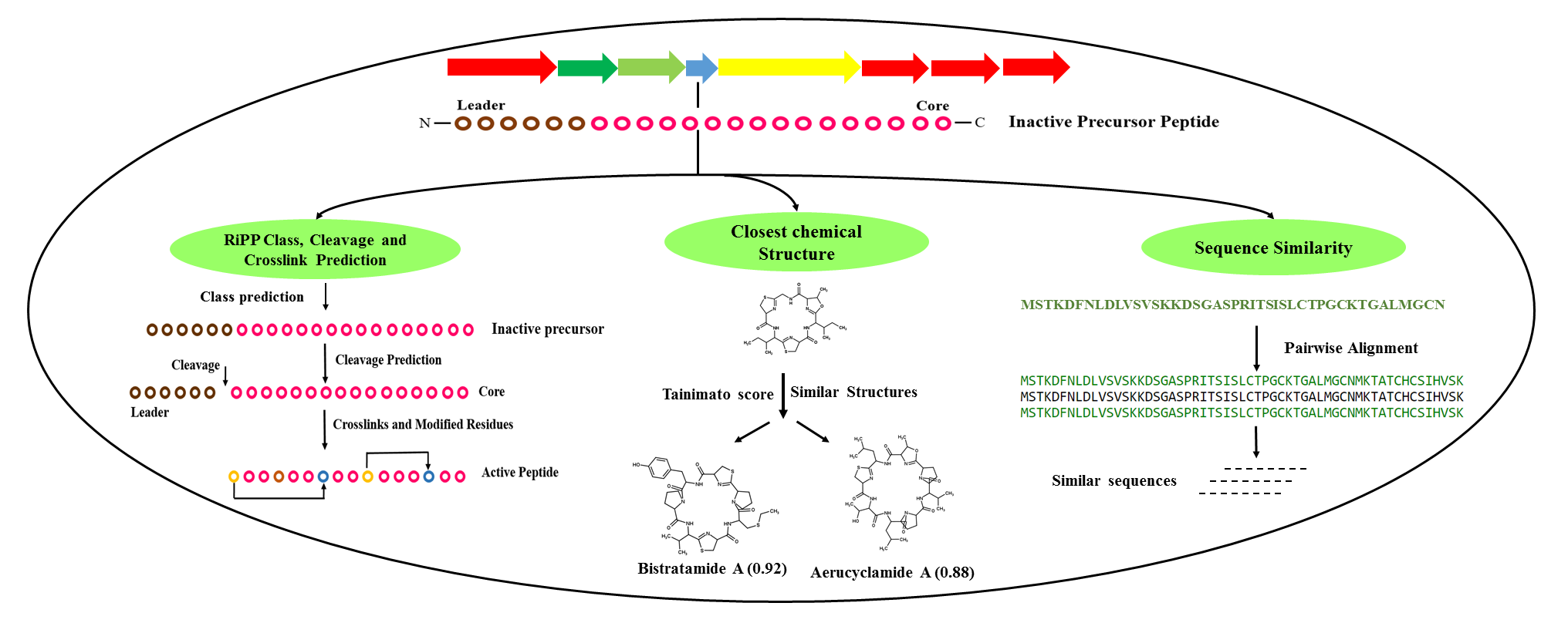
RiPPMiner is a machine learning based webserver for deciphering chemical structures of Ribosomally synthesized Post translationally modified Peptides (RiPPs) which constitute one of the major class of pharmaceutically important antimicrobial compounds (e.g. NisinA). It uses amino acid sequence of the precursor polypeptide of RiPPs as input.
Tools: RIPPMiner uses SVM to classify the precursor into nine sub-classes of RiPPs. For classes like Lanthipeptide, Cyanobactin and Lassopeptide, RiPPMiner predicts the leader cleavage site and finally complex crosslinking and post-translationally modified residues are predicted in the core peptide. For Thiopeptide it predicts crosslinking and modified residues. RiPPMiner can identify correct PTM and crosslink pattern in a query RiPP core peptide from among a very large number of combinatorial possibilities using machine learning. RiPPMiner also provides GUI for visualization of the predicted chemical structure and also search for characterized RiPPs similar to a given peptide sequence or a given chemical structure.
Database: RiPPMiner derives its predictive power from training using a manually curated database of more than 500+ experimentally characterized RiPPs belonging to 13 Subclasses. These Classes include Lanthipeptide, Bottromycin, Cyanobactin, Glycocin, Lassopeptide, Linaridin, Linearazol, Microcin, Sactipeptide, Thiopeptide, Auto Inducing Peptide, ComX, Bacterial Head to Tail Cyclized. Each RiPP has information like Modification System, Complete Sequence, Leader and Core Sequence, modified residues, Cross-links, Pubmed, PDB ID, Gene Cluster among others.


Visitors:

Tools: RIPPMiner uses SVM to classify the precursor into nine sub-classes of RiPPs. For classes like Lanthipeptide, Cyanobactin and Lassopeptide, RiPPMiner predicts the leader cleavage site and finally complex crosslinking and post-translationally modified residues are predicted in the core peptide. For Thiopeptide it predicts crosslinking and modified residues. RiPPMiner can identify correct PTM and crosslink pattern in a query RiPP core peptide from among a very large number of combinatorial possibilities using machine learning. RiPPMiner also provides GUI for visualization of the predicted chemical structure and also search for characterized RiPPs similar to a given peptide sequence or a given chemical structure.
Database: RiPPMiner derives its predictive power from training using a manually curated database of more than 500+ experimentally characterized RiPPs belonging to 13 Subclasses. These Classes include Lanthipeptide, Bottromycin, Cyanobactin, Glycocin, Lassopeptide, Linaridin, Linearazol, Microcin, Sactipeptide, Thiopeptide, Auto Inducing Peptide, ComX, Bacterial Head to Tail Cyclized. Each RiPP has information like Modification System, Complete Sequence, Leader and Core Sequence, modified residues, Cross-links, Pubmed, PDB ID, Gene Cluster among others.