Tool: There are following four facilities are available under Tool Section:
1. RiPP Class Prediction
2. RiPP Cleavage & Crosslinks Prediction
3. Sequence Similarity Search
4. Closest Structure Search
1. RiPP Class Prediction
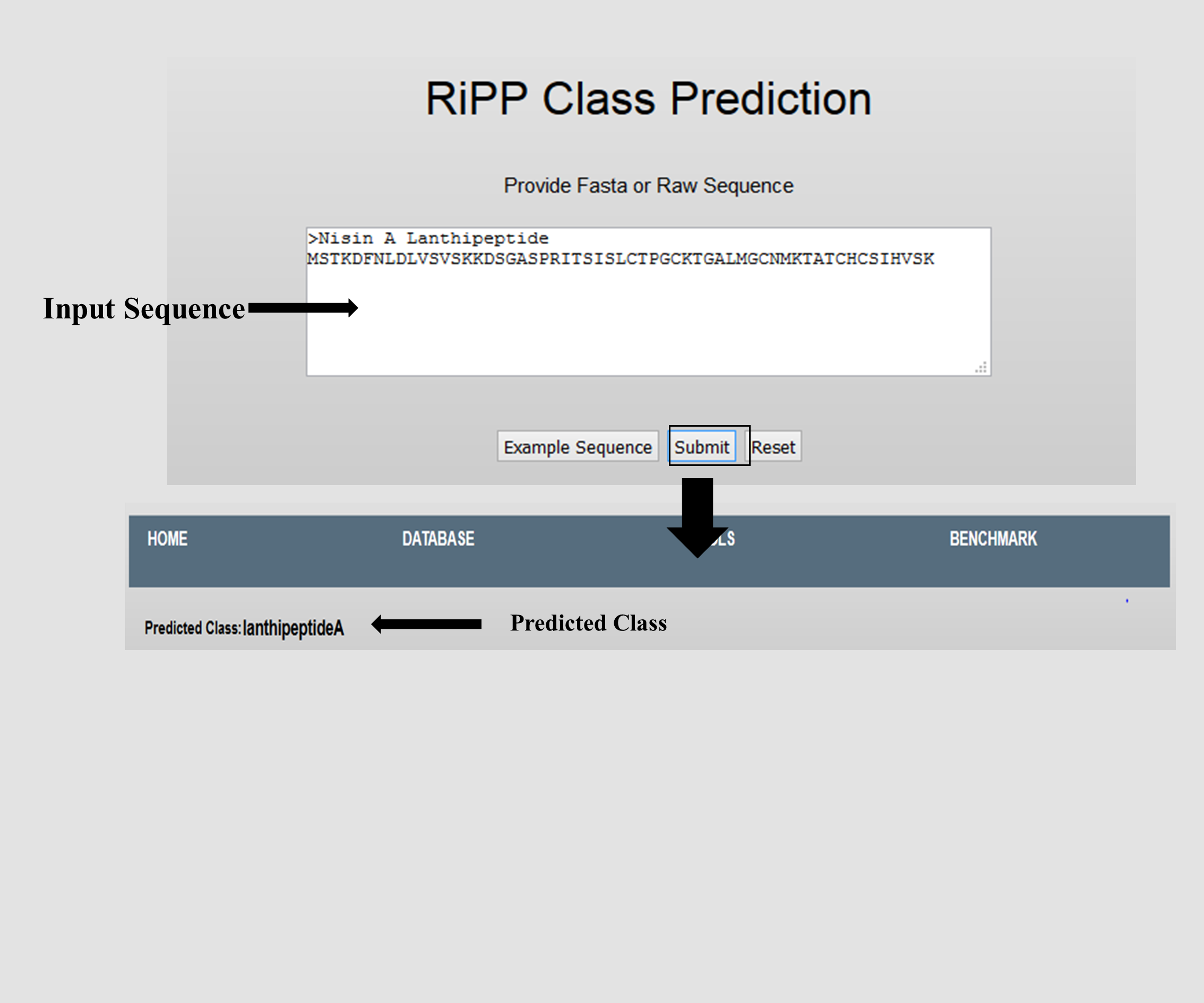
This SVM based classifier can be used to predict the 7 Classes of RiPP peptides.
These seven classes are: Auto Inducing peptide, Bacterial head-to-tail cyclized peptide, Lanthipeptide, ComX, Cyanobactin, Lassopeptide, Procholorosin, Sactipeptide, Thiopeptide.
It can also predict the three major subclasses of Lanthipeptides (i.e. Lanthipeptides A, B and C).
2. RiPP Cleavage & Crosslinks Prediction
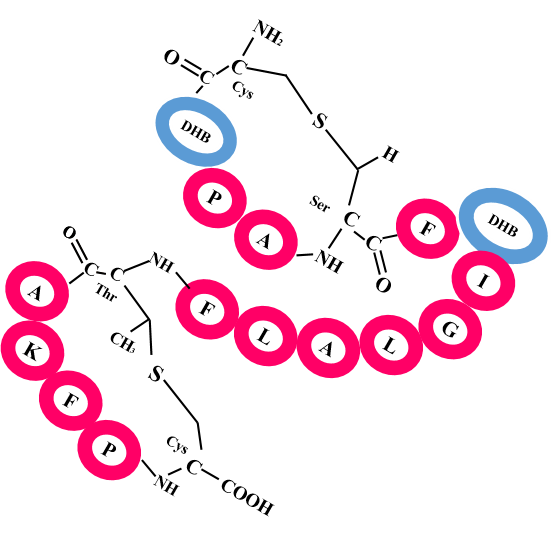
A. Lanthipeptide
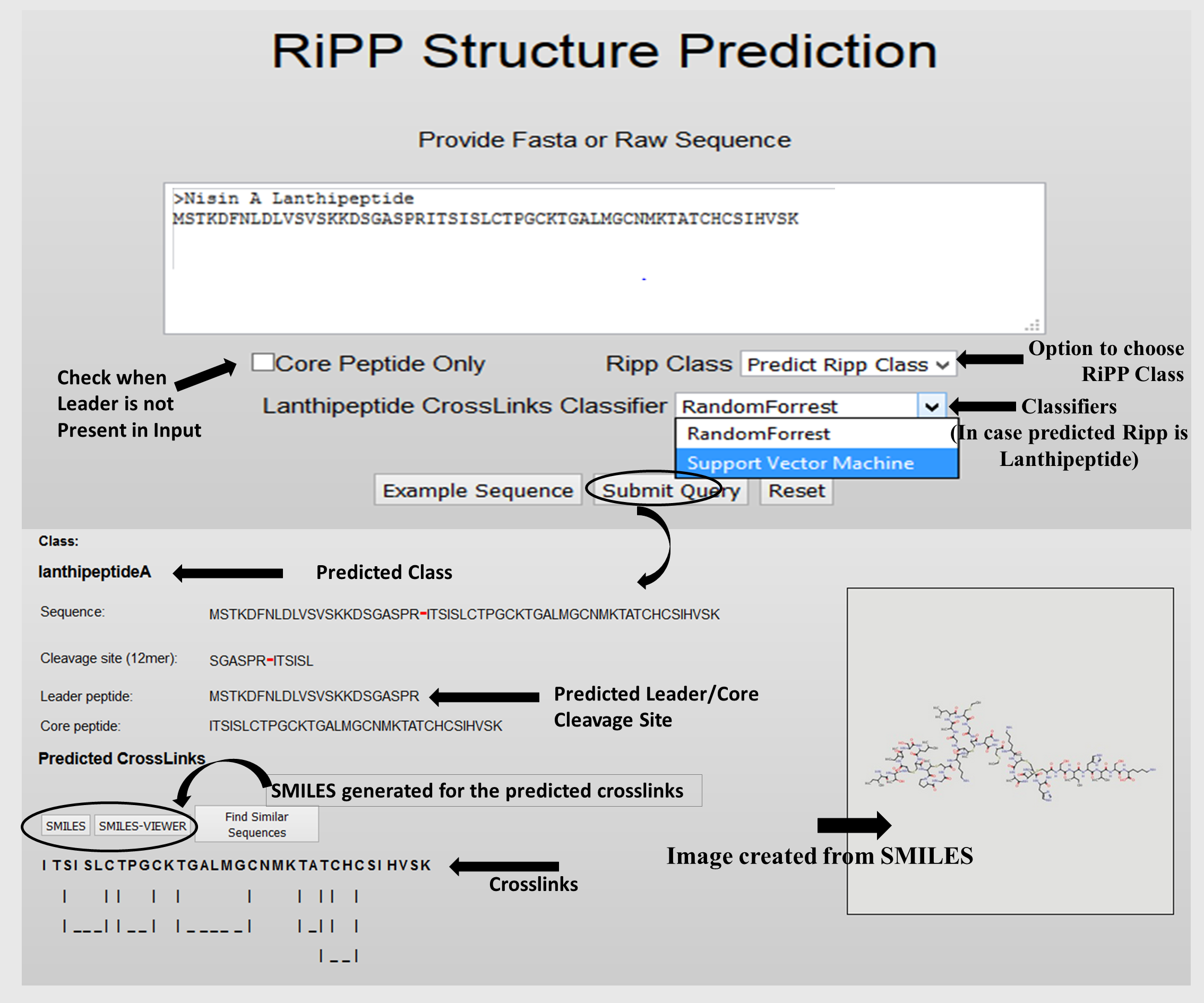
The Classifier first predict the Class of Input RiPP sequence, if the predicted class is any of the these three classes, Lanthipeptide, Lassopeptide, or Cyanobactin, then further prediction of Leader peptide Cleavage site and cross-links within core peptide is carried out.
B. Lassopeptide
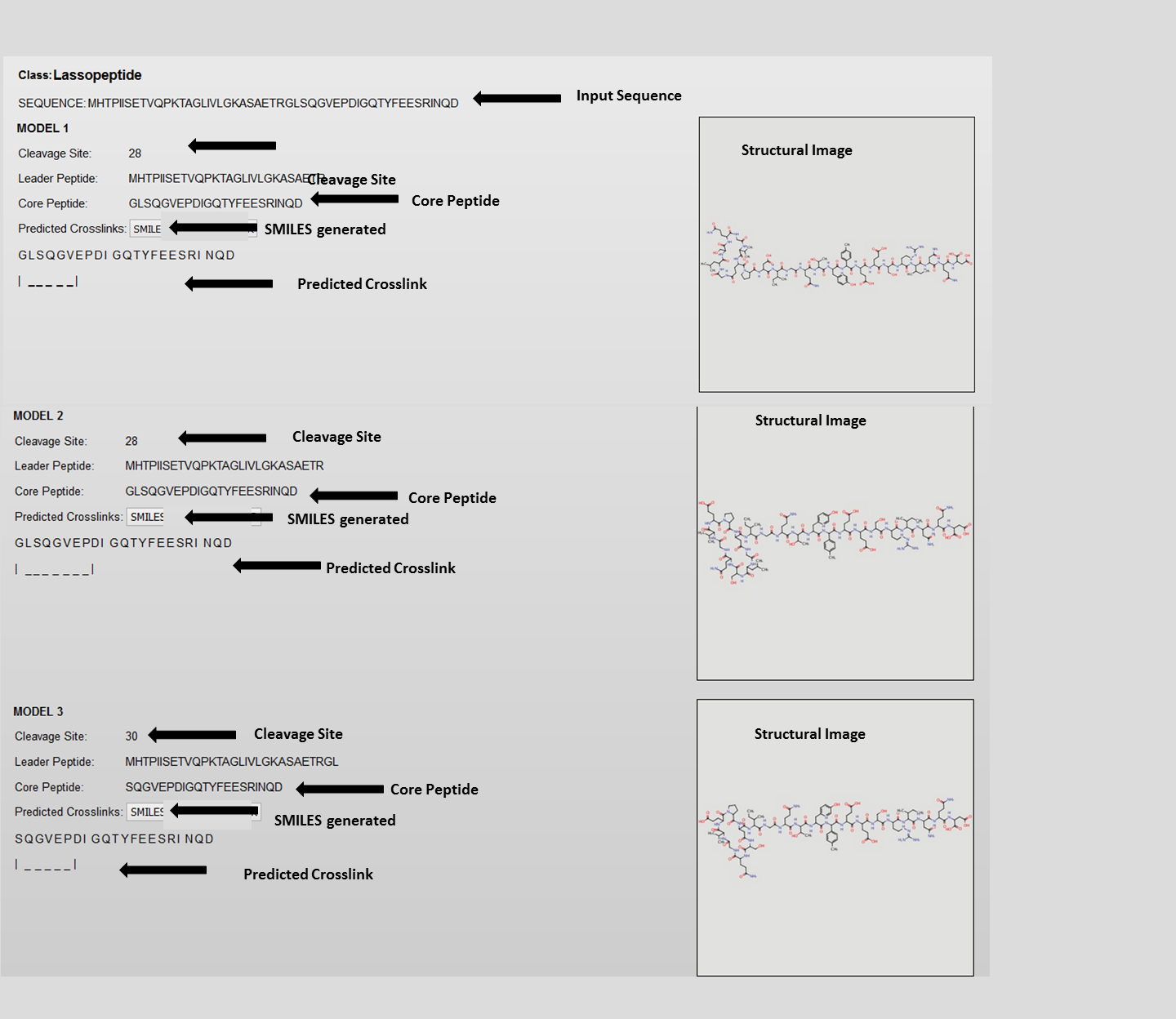
C. Cyanobactin

3. Sequence Similarity Search
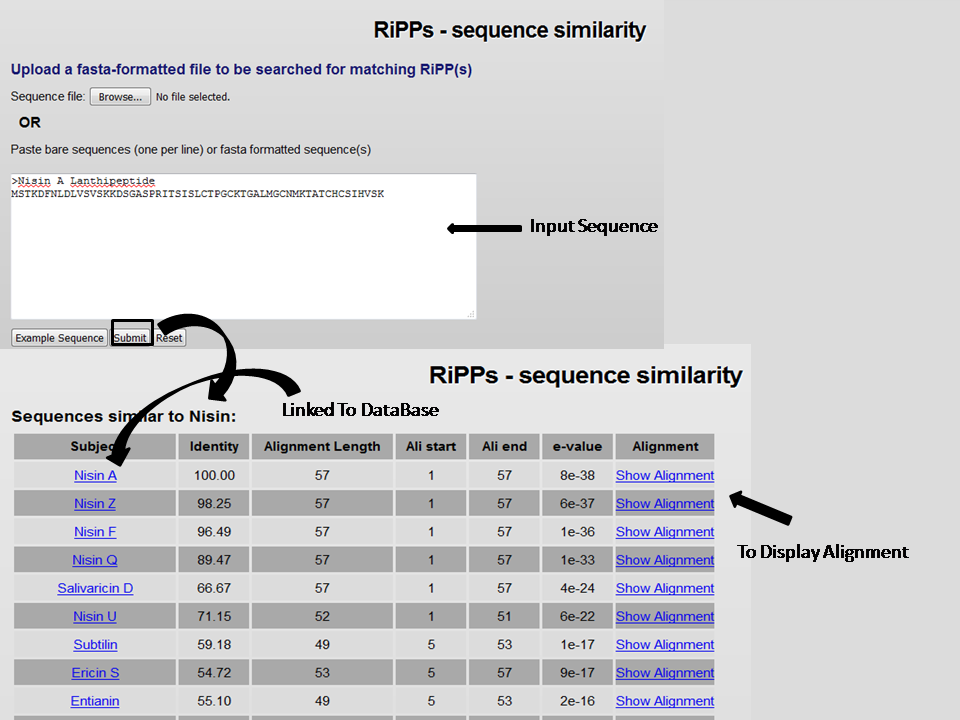
It is a Blastp based tool where one can do the protein sequence based similiraity search to find the closest neighbors of RiPP peptides.
4. Closest Structure Search
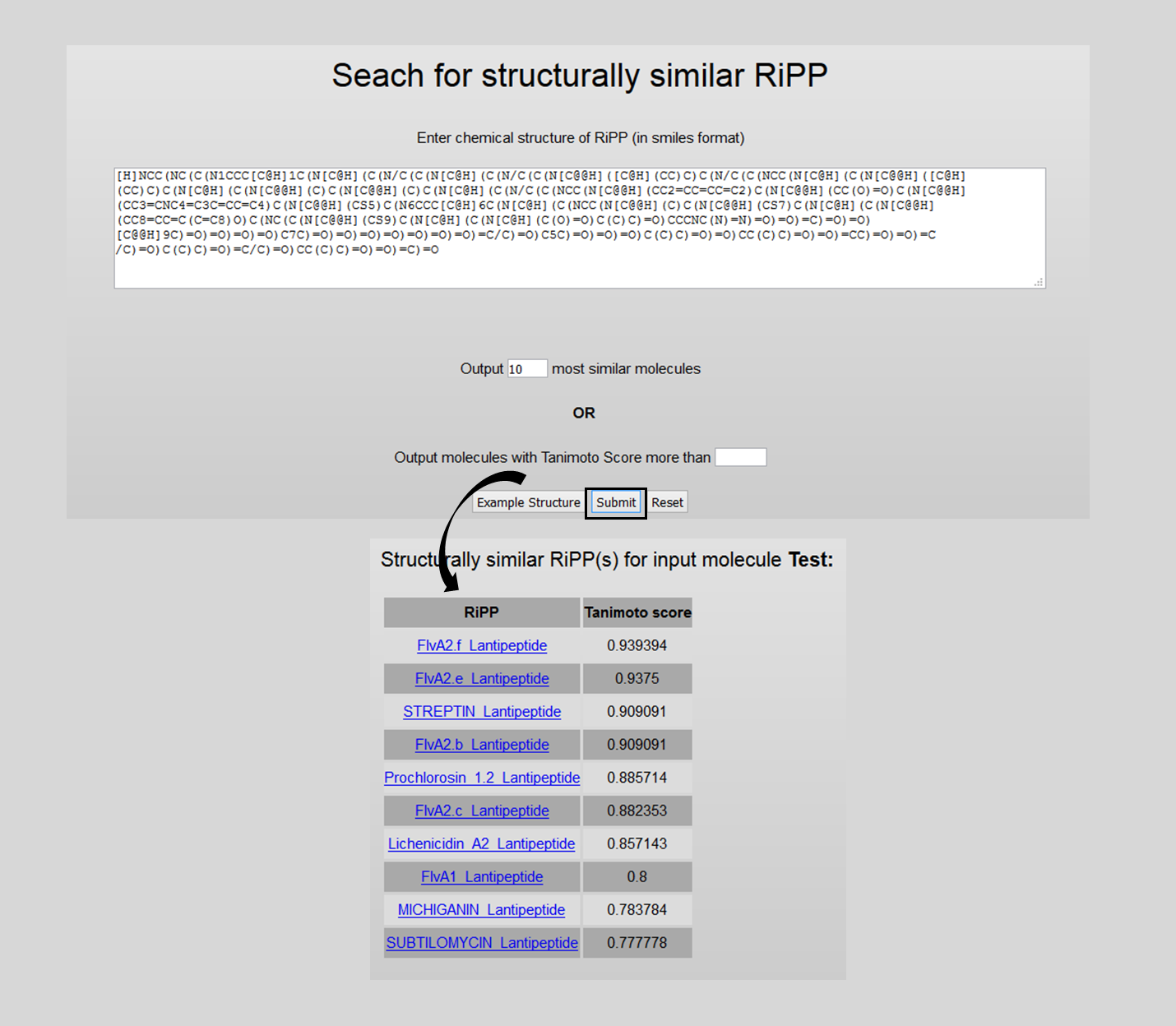
Here, one can do SMILES based search to find closest RiPP structures. The number of outputs can be restrcited by either providing an Integer 'n' (Top 'n' outputs will be shown; default is '10') or by giving a Tanimoto Score cutoff.
5. Database
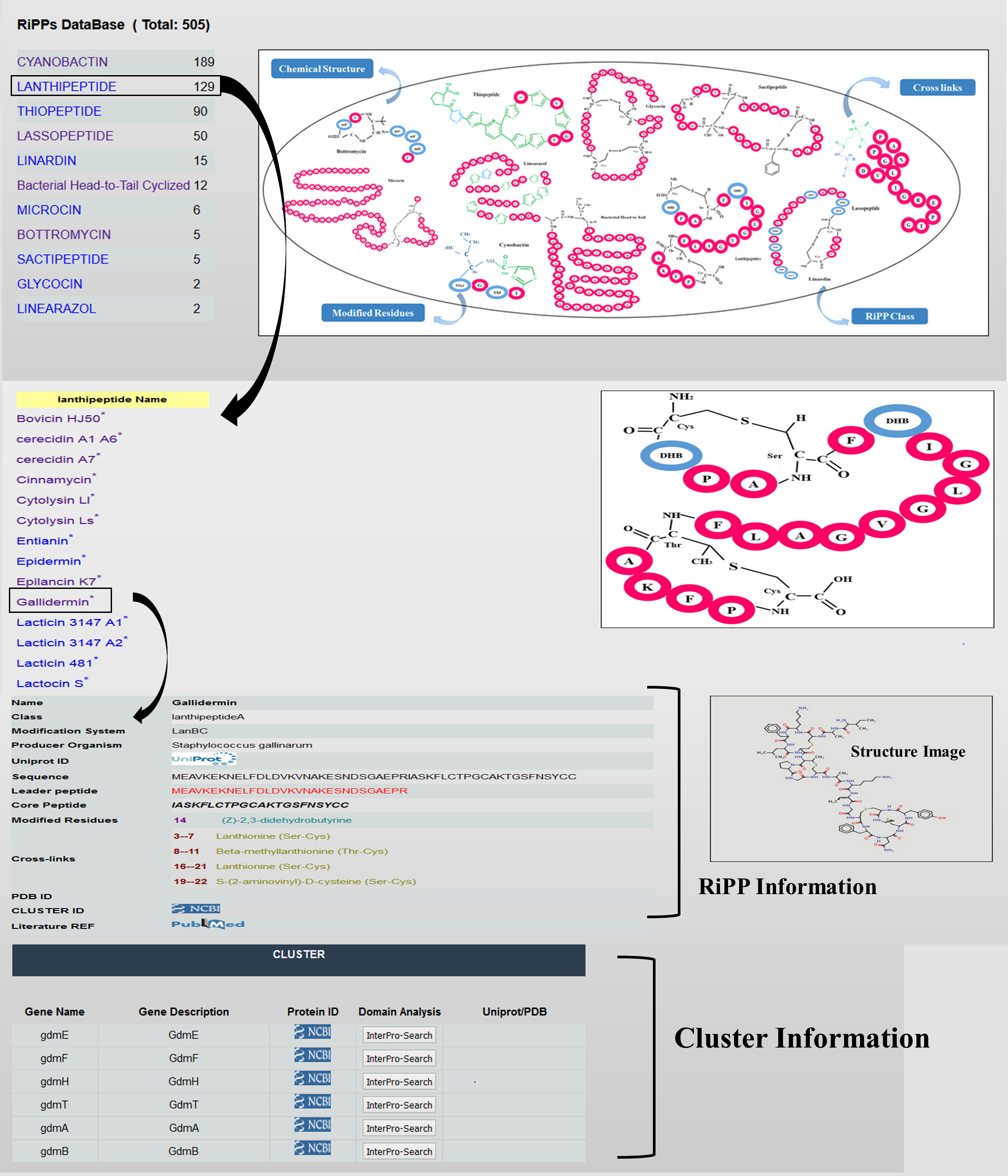
The database part currently holds 500+ manually curated peptides belonging to 11 different RiPP Classes.